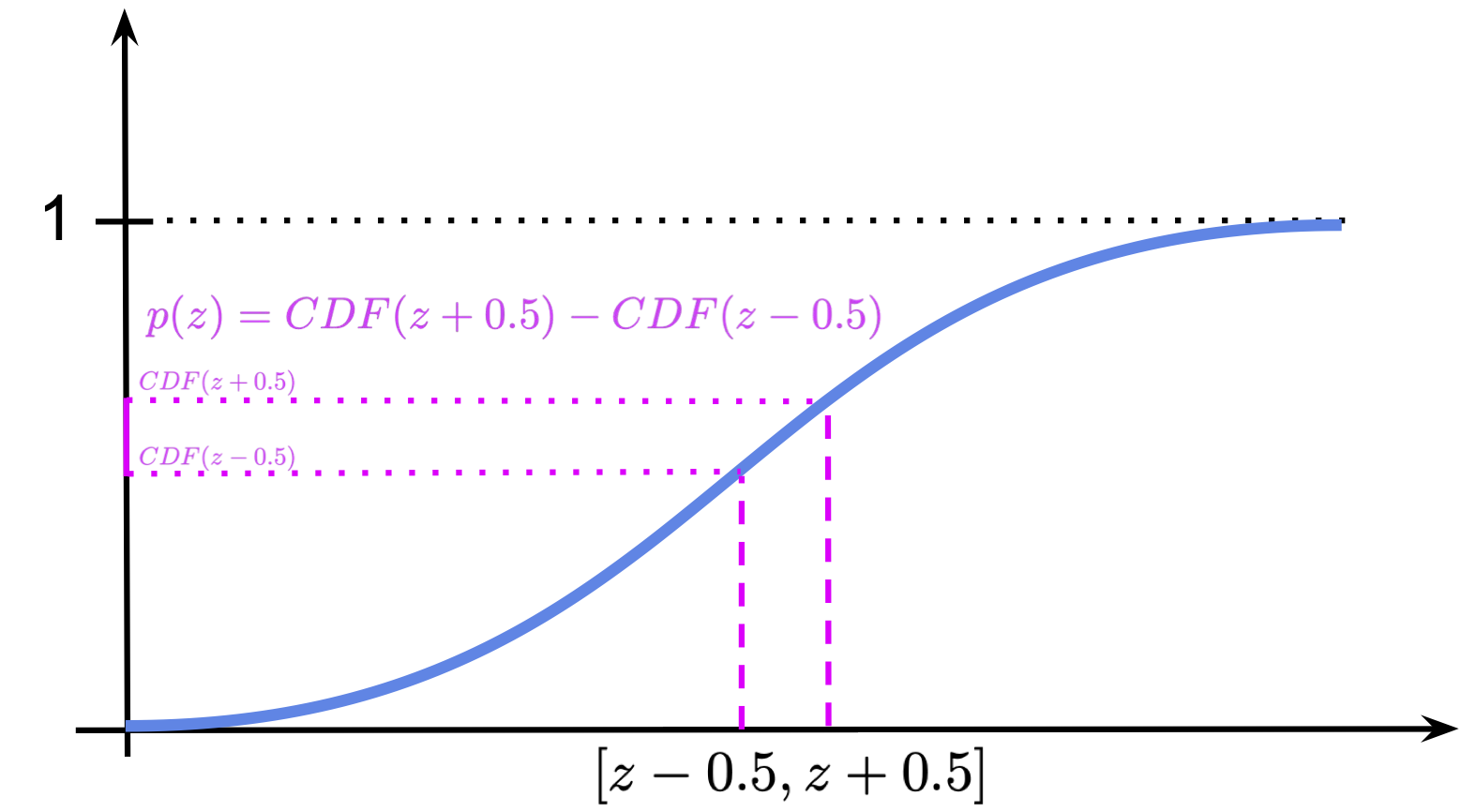
About

Jakub M. Tomczak, Ph.D.
- CV: [PDF]
- Website: jmtomczak.github.io
- Email: jmk.tomczak
 gmail.com
gmail.com - City: San Francisco, USA
A Generative AI Leader with 15+ years of experience in machine learning, deep learning, and Generative AI. Proven track of leading research projects (academic: 50+ AI MSc&PhD, 10+ industrial: Computer Vision, LLMs, Foundation Models, Agentic AI), carrying out cutting-edge research (2 patents & 1 application, 20+ conference papers: NeurIPS, ICML, ICLR, AISTATS, UAI, CVPR, ICCV, 25+ journal papers), and securing funds (EUR 2,3M). Experienced in and enjoying managing people (3y in the industry, 8y in academia). Effective team leader encouraging initiative & independence, and facilitating cross-functional collaboration, serving as a (fractional) AI leader (head/director/CTO) for companies (e.g., eBay, Qualcomm) and startups. A Program Chair of NeurIPS 2024 (the largest AI conference so far), and the author of the first fully comphrehensive book on GenAI ("Deep Generative Modeling"). He is also the founder of Amsterdam AI Solutions.
Facts
Years of Experience
Supervised projects
Top publications
Funding gathered (EUR)
Additional Information
Presentations
My presentations at conferences, workshops, for academic and industrial research groups, summer schools
DeeBMed
My project that was carried out from 2016/10/1 to 2018/09/30 within the Marie Skłodowska-Curie Action (Individual Fellowship), financed by the European Comission.
My Book
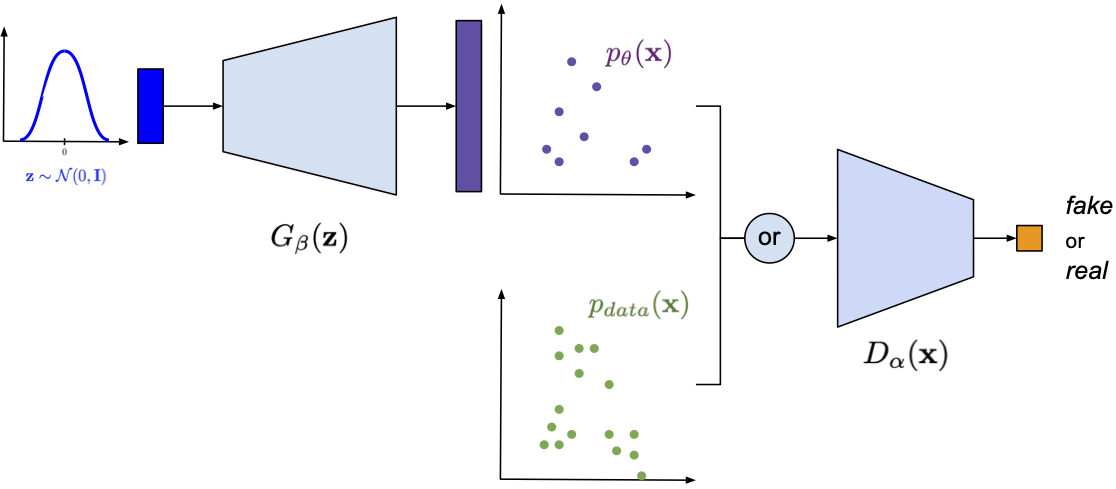
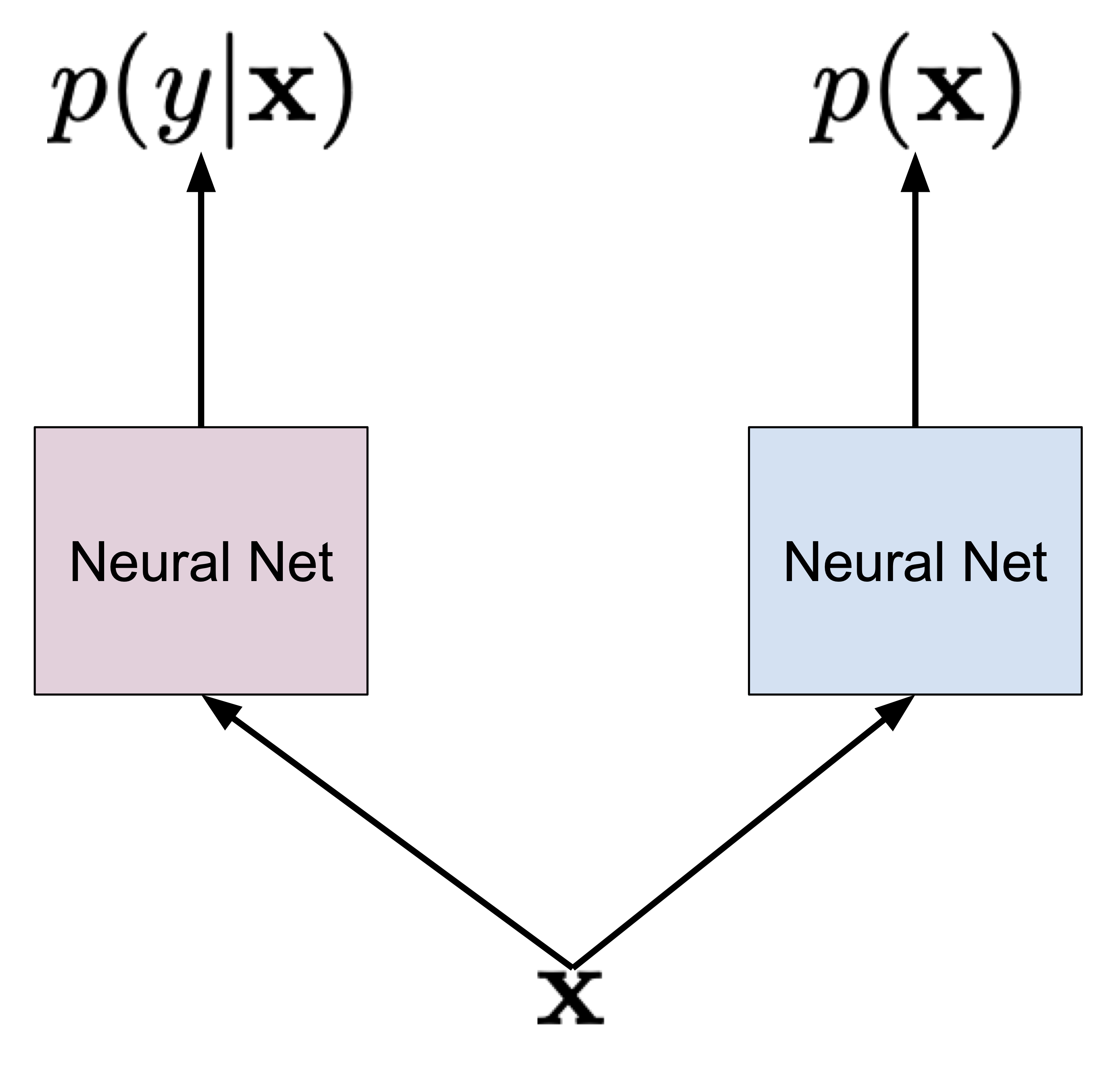
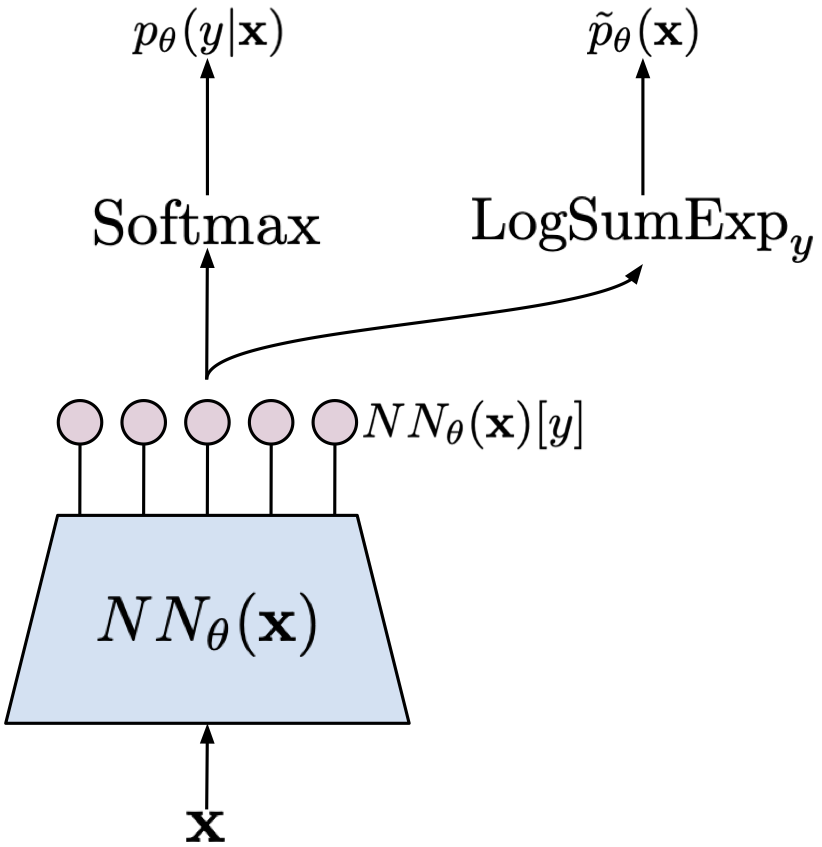
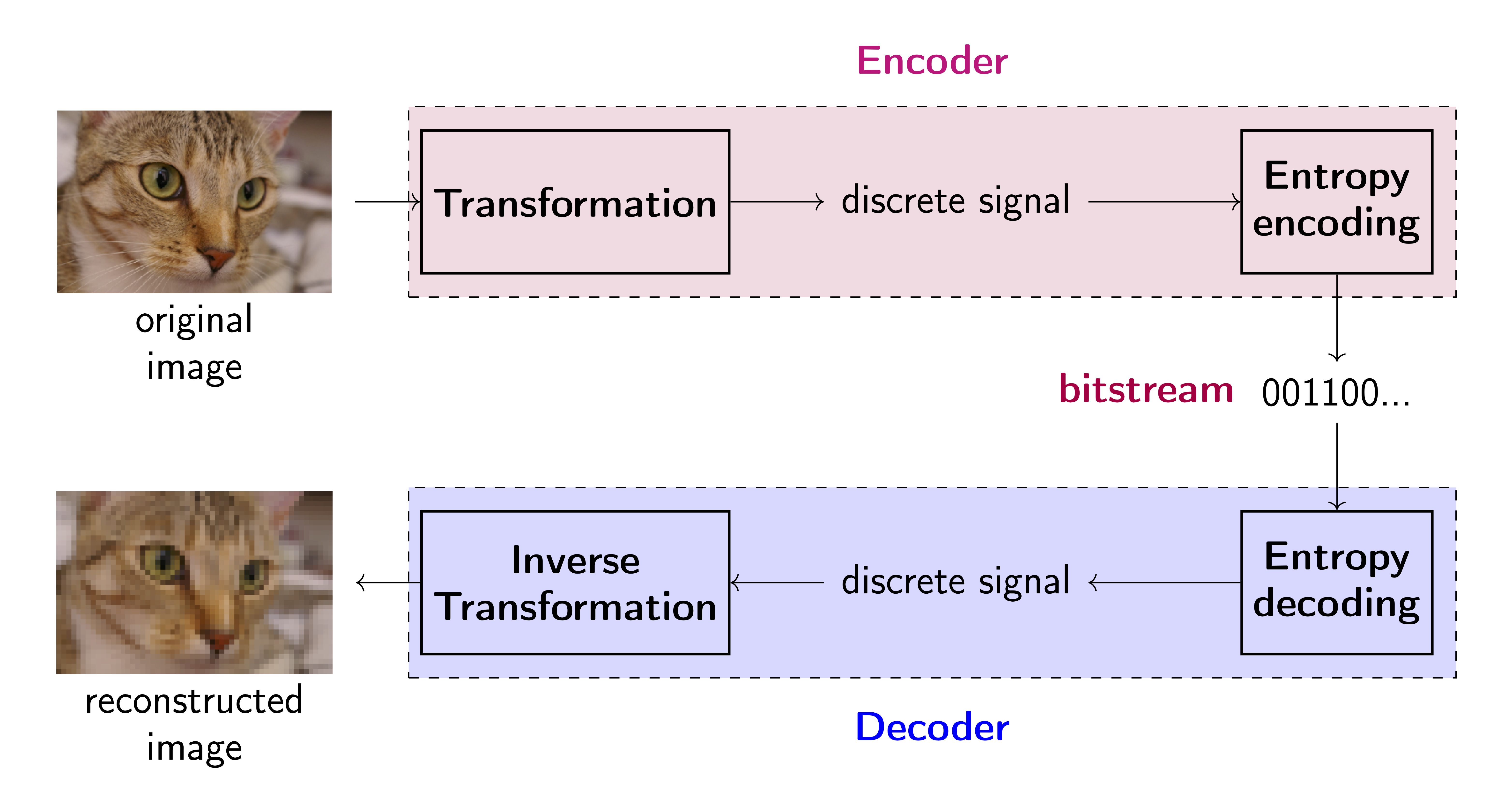
My book on Generative Artificial Intelligence. This is the first comprehensive book on deep generative modeling explaining theory behind Diffusion-based models, Variational Auto-Encoders, GANs, autoregressive models, flow-based models, and energy-based models. Moreover, each chapter is associated with code snippets presenting how the beforementioned models could be implemented in PyTorch.
Resume
Education
Ph.D. in Computer Science/Machine Learning (with honors)
Mar 2013
Wroclaw University of Technology, Poland
Title: Incremental Knowledge Extraction from Data for Non-Stationary Objects
Supervisor: Prof. Jerzy Swiatek
M.Sc. in Computer Science
Dec 2009
Blekinge Institute of Technology, Sweden
M.Sc. in Computer Science (the best thesis in Poland)
Jul 2009
Wroclaw University of Technology, Poland
Work Experience
Senior Staff Research Scientist, Generative AI x Science
Dec 2024 - Present
Chana Zuckerberg Initiative, Bay Area, the USA
Group Leader & Associate Professor, Generative AI
Feb 2023 - Jan 2025
Eindhoven University of Technology, Eindhoven, the Netherlands
Founder, Fractional Generative AI leadership
Oct 2022 - Present
Amsterdam AI Solutions, Amsterdam, the Netherlands
Head AI Advisor, Generative AI for Drug Discovery
Mar 2022 - Present
NatInLab, Amsterdam, the Netherlands
Group Co-Leader & Assistant Professor, Generative AI
Nov 2019 - Feb 2023
Vrije Unviersiteit Amsterdam, the Netherlands
Staff Engineer (Deep Learning Scientist)
Oct 2018 - Dec 2019
Qualcomm AI Research, Amsterdam, the Netherlands
Principal Investigator (Marie Sklodowska-Curie Individual Fellow), Generative AI
Oct 2016 - Sept 2018
Universiteit van Amsterdam, the Netherlands
Senior Scientist, AI for Drug Discovery
Feb 2016 - Jun 2016
INDATA SA, Wroclaw. Poland
Assistant Professor, Machine Learning
Oct 2014 - Sept 2016
Wroclaw University of Technology. Poland
Postdoc, Machine Learning
Oct 2012 - Sept 2014
Wroclaw University of Technology. Poland
Ph.D. student / Research assistant, Machine Learning
Jun 2009 - Sept 2012
Wroclaw University of Technology. Poland
Publications
Book
J.M. Tomczak, ''Deep Generative Modeling'', Springer, Cham, 2022. [More]
Full list of papers
Please check my ![]() Google Scholar
Google Scholar
BLOG
- All
- Intro
- ARMs
- Flows
- VAEs
- DBGMs
- SBGMs
- GANs
- Joint
- Applications
Contact
Location:
Amsterdam, the Netherlands
Email:
jmk.tomczak![]() gmail.com
gmail.com